graph_clustering <- function(W, k, nu, td) {
N <- nrow(W)
# Step 1: Compute the parametrized random walk operator P(ν)
D <- diag(rowSums(W))
P_nu <- solve(D) %*% W %*% diag(nu)
# Step 2: Compute the parametrized random walk diffusion kernel K(td,ν)
K_td_nu <- expm::expm(td * (P_nu - diag(N)))
# Step 3: Apply k-means clustering to the rows of K(td,ν)
kmeans_result <- kmeans(K_td_nu, centers = k)
# Step 4: Obtain the k-partition Vtd based on the clustering result
Vtd <- kmeans_result$cluster
# Step 5: Return the partition
return(Vtd)
}Claude helps to create an R package
Parametrized Random Walk Diffusion Kernel Clustering
Reinhold Koch
2024-09-04
Introduction
Based on arxiv.org/pdf/2210.00310 I asked Claude, to help me create an R package that implements the described algorithms. The result is accessible in github.com/baselDataScience/prwdkc.
My first encounter with Claude!
The Algorithm in the Paper
I said: “write me a R function that implements the following algorithm:” 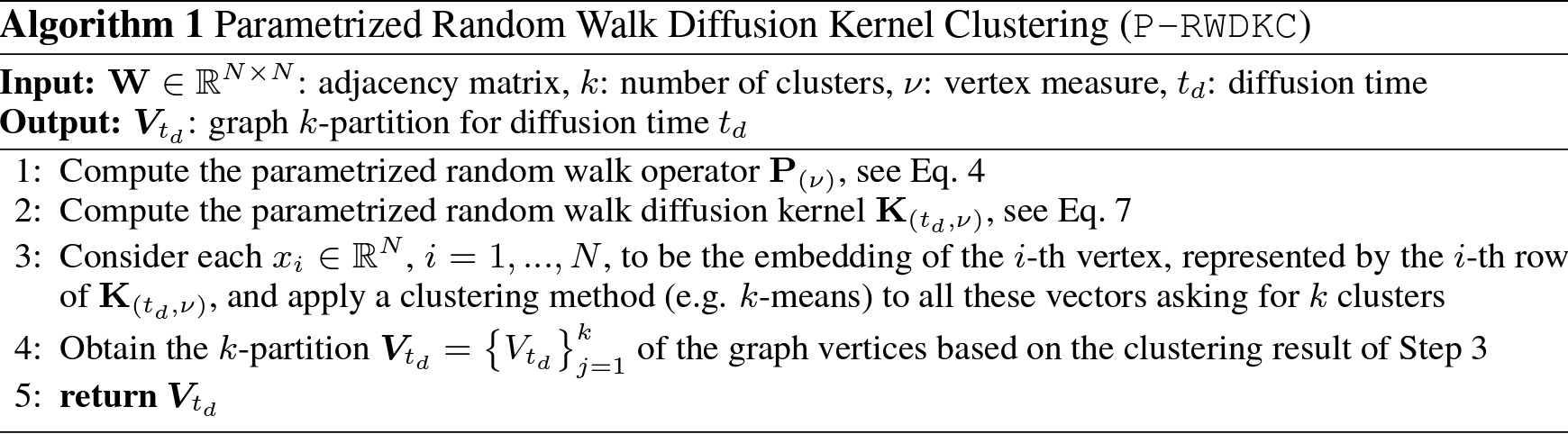
Claude’s first attempt:
My Impression
- Claude always answers very politely (later): “I apologize for the confusion. You’re right, and I appreciate your correction.”
- Claude can produce R code
- Claude knows some matrix functions
- Claude knows about package expm
- but
- why should K_td_nu be an exponential matrix??
“please add input checks”
# Input checks
if (!is.matrix(W) || !isSymmetric(W) || any(W < 0)) {
stop("W must be a symmetric matrix with non-negative entries")
}
if (!is.numeric(k) || length(k) != 1 || k <= 0 || k != round(k)) {
stop("k must be a positive integer")
}
if (!is.numeric(nu) || length(nu) != nrow(W) || any(nu <= 0)) {
stop("nu must be a positive numeric vector with length equal to the number of rows in W")
}
if (!is.numeric(td) || length(td) != 1 || td <= 0) {
stop("td must be a positive number")
}W needs not be symmetric though …
Interaction with Claude
- certainly helpful to get going
- deals reasonably well with mathematical notations like subscripts or exponents
- but can be strenuous to nudge him in the right direction
back to clustering
- usable code is now in github.com/baselDataScience/prwdkc
- which functions to provide?
- some helpful examples should be added …
- and some tests!
spirals - what can prwdkc do?
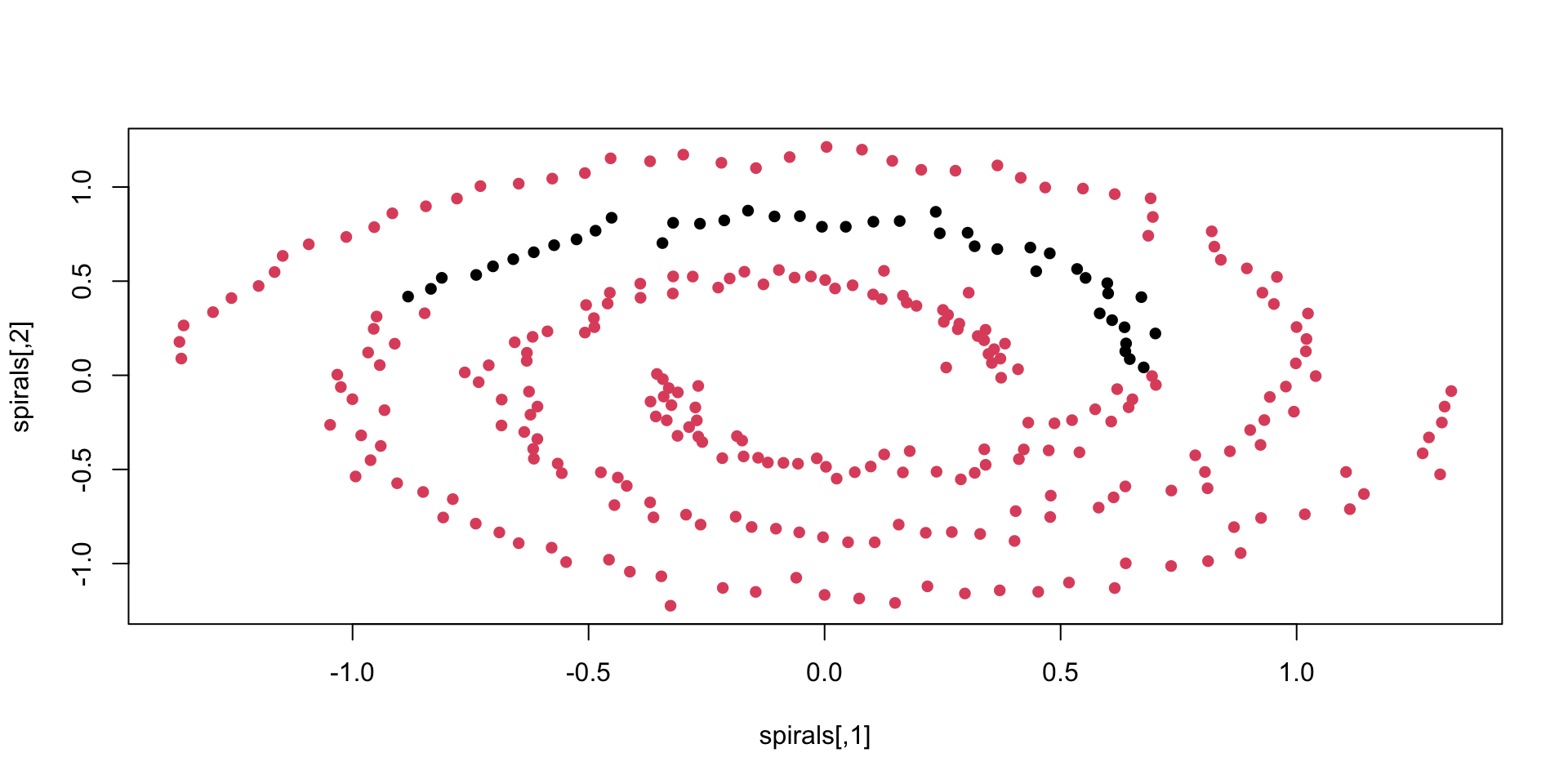
spirals - with kernlab
not too great - see for comparison spectral clustering:
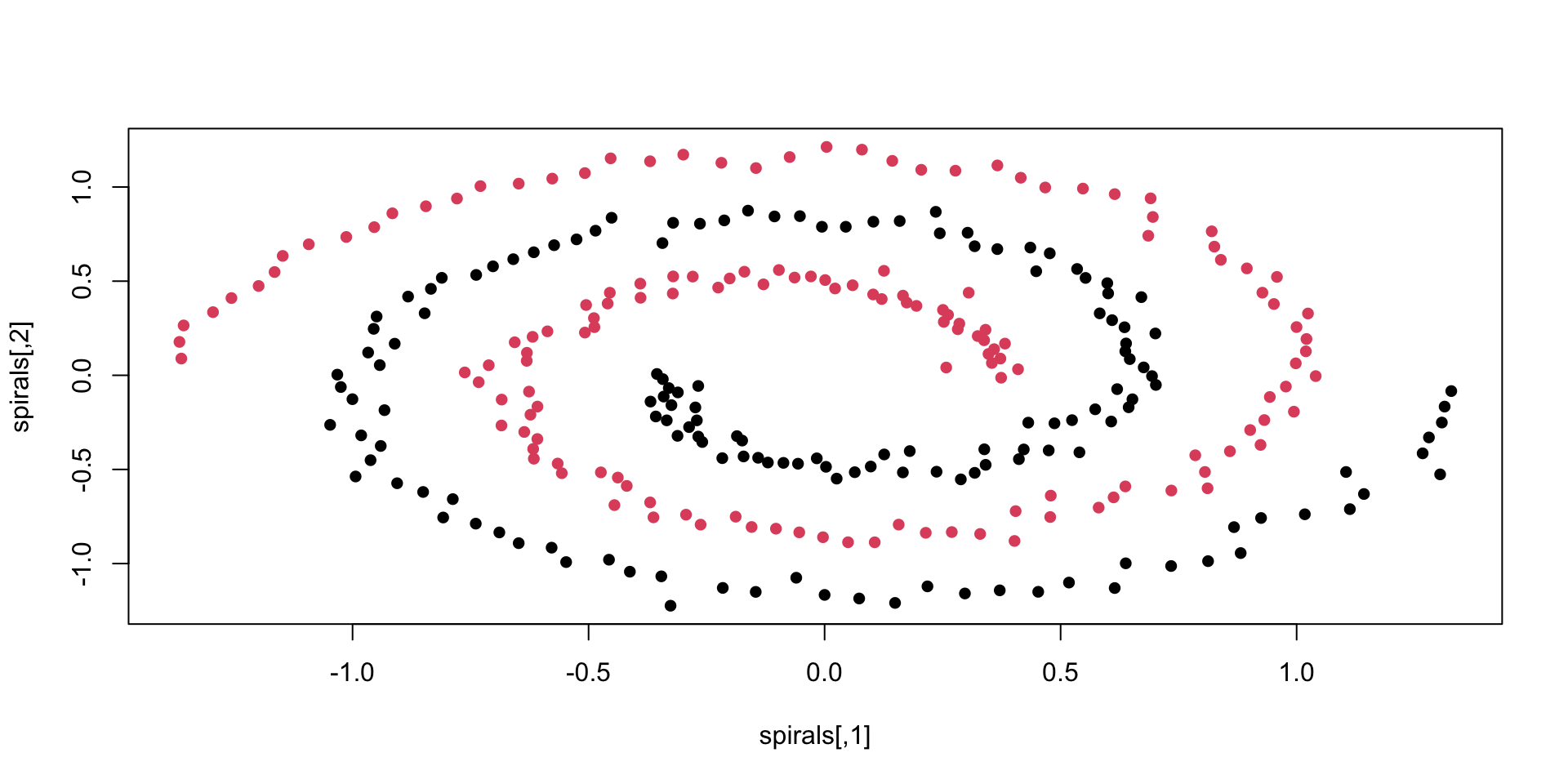
other considerations
- prwdkc does do better with other test cases and with examples from the Machine Learning Repository https://archive.ics.uci.edu/datasets
- sometimes no embedding in Rn is given, only a graph with adjacency matrix - for instance links between web pages
- first(?) algorithm able to deal with directed graphs,
i.e. non-symmetric adjacency matrices
what is the idea behind prwdkc?
“diffusion” along a graph: 
where to start “diffusion”?
- heuristics so far,
trying different start parameter nu in prwdkc()
when to stop “diffusion”?
- implemented via the optstop() function
- dependent on the eigenvalues of the adjacency matrix
source in https://github.com/BaselDataScience/baseldatascience.github.io/blob/main/posts/prwkdc.qmd